An American Non-profit organization called Monterey Bay Aquarium Research Institute (MBARI) has been able to join two different technologies to make the perfect tool to study the Environmental DNA (eDNA) of marine species using scientists using autonomous underwater robots to sample and it has been proving to be a great addition to the environmental sciences so far. Environmental DNA (eDNA) is an emerging and robust method for use in marine research, conservation, and management, yet time- and resource-intensive protocols limit the scale of implementation.
eDNA also allows scientists to detect the presence of aquatic species from the tiny bits of genetic material they leave behind. This DNA soup offers clues about biodiversity changes in sensitive areas, the presence of rare or endangered species, and the spread of invasive species—all critical to understanding, promoting, and maintaining a healthy ocean. This tool helps the scientists to collect the eDNA with absolute ease so that they can focus on the research and discovery part rather than finding the sample in the first place.
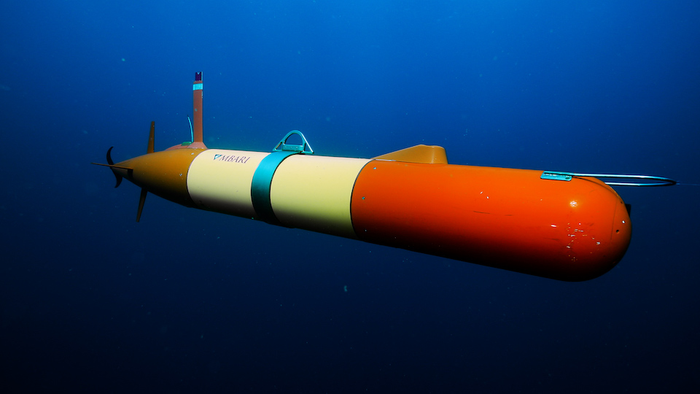
In a significant step forward for monitoring the biodiversity of marine systems, a new study by MBARI scientists is using autonomous underwater robots to sample environmental DNA (eDNA). They combined two novel independent platforms: the long-range autonomous underwater vehicle (LRAUV) and the Environmental Sample Processor (ESP).
By combining both technologies, scientists can expand the scale of ocean monitoring over time and space. Technology innovations like this are revolutionizing ocean conservation efforts. Both technologies allowed scientists to maintain a persistent presence in the ocean and monitor changes in sensitive ecosystems in ways that were not possible previously.
A co-author of the study and collaborator at the National Oceanic and Atmospheric Administration (NOAA), Kelly Goodwin, said;
“Organisms move as conditions change in our oceans and Great Lakes, affecting the people and economies that rely on those species. We need cheaper and more nimble approaches to monitor biodiversity on a large scale. This study provides the synergistic development of eDNA and uncrewed technologies we need in direct response to priorities laid out in the NOAA Omics Strategic Plan.”
These scientists collaborated with the NOAA Atlantic Oceanographic and Meteorological Laboratory and the University of Washington, scientists completed three expeditions in the Monterey Bay National Marine Sanctuary. The team coordinated sample collection between MBARI’s three research vessels, the NOAA Fisheries ship Reuben Lasker, and a fleet of MBARI’s LRAUVs.
Scientists lowered collectors of the robots to a certain depth to gather and preserve water samples to collect and preserve water samples. In the meantime, an LRAUV equipped with an ESP sampled and preserved eDNA at similar depths and locations. The eDNA samples were sent back to the lab for further analysis.
Using a technique called metabarcoding, scientists analyzed eDNA samples and translated the data into a measure of biodiversity. They found four different types of gene markers, each representing a slightly different level of the food web. The combined data provided a more comprehensive picture of community composition. Similar biodiversity patterns were seen in samples taken from research ships and autonomous vehicles.
Last month, MBARI tweeted a video of the working of its Autonomous Robot and said;
“A long-range autonomous underwater vehicle is a nimble robot that can travel to remote areas of the ocean for extended missions. Autonomous technology allows MBARI researchers to maintain a persistent presence in the ocean and collect data that informs effective management.”
Kobun Truelove, a biological oceanographer at MBARI and the lead author of the paper said in his interview about the recent progression of the project;
“We know that eDNA is an incredibly powerful tool for studying ocean communities, but we’ve been limited by what we can accomplish using crewed research vessels. Now, autonomous technology is helping us make better use of our time and resources to study new parts of the ocean, The findings from the study mark an exciting step forward for monitoring marine ecosystems. This work is all about increasing the scale of eDNA research. Instead of looking at an individual species, we can start to characterize biological community structure in the ocean more broadly.”
The specialty of this method is that it looks for short DNA excerpts and provides a breakdown of the groups present in the sample. This technique is especially helpful for translating eDNA data into a measure of biodiversity. The researchers analyzed four different types of gene markers, each representing a slightly different level of the food web. All the data collected and analyzed via different spectrums of intrigue can develop better imagery of the aqueous history that humans have ever been able to estimate.
Read More: